Running a parallel job
Last updated on 2025-03-06 | Edit this page
Overview
Questions
- “How do we execute a task in parallel?”
- “What benefits arise from parallel execution?”
- “What are the limits of gains from execution in parallel?”
Objectives
- “Construct a program that can execute in parallel.”
- “Prepare a job submission script for the parallel executable.”
- “Launch jobs with parallel execution.”
- “Record and summarize the timing and accuracy of jobs.”
- “Describe the relationship between job parallelism and performance.”
We now have the tools we need to run a multi-processor job. This is a very important aspect of HPC systems, as parallelism is one of the primary tools we have to improve the performance of computational tasks.
Our example implements a method for estimating the value of π, the
ratio of the circumference to the diameter of a circle.
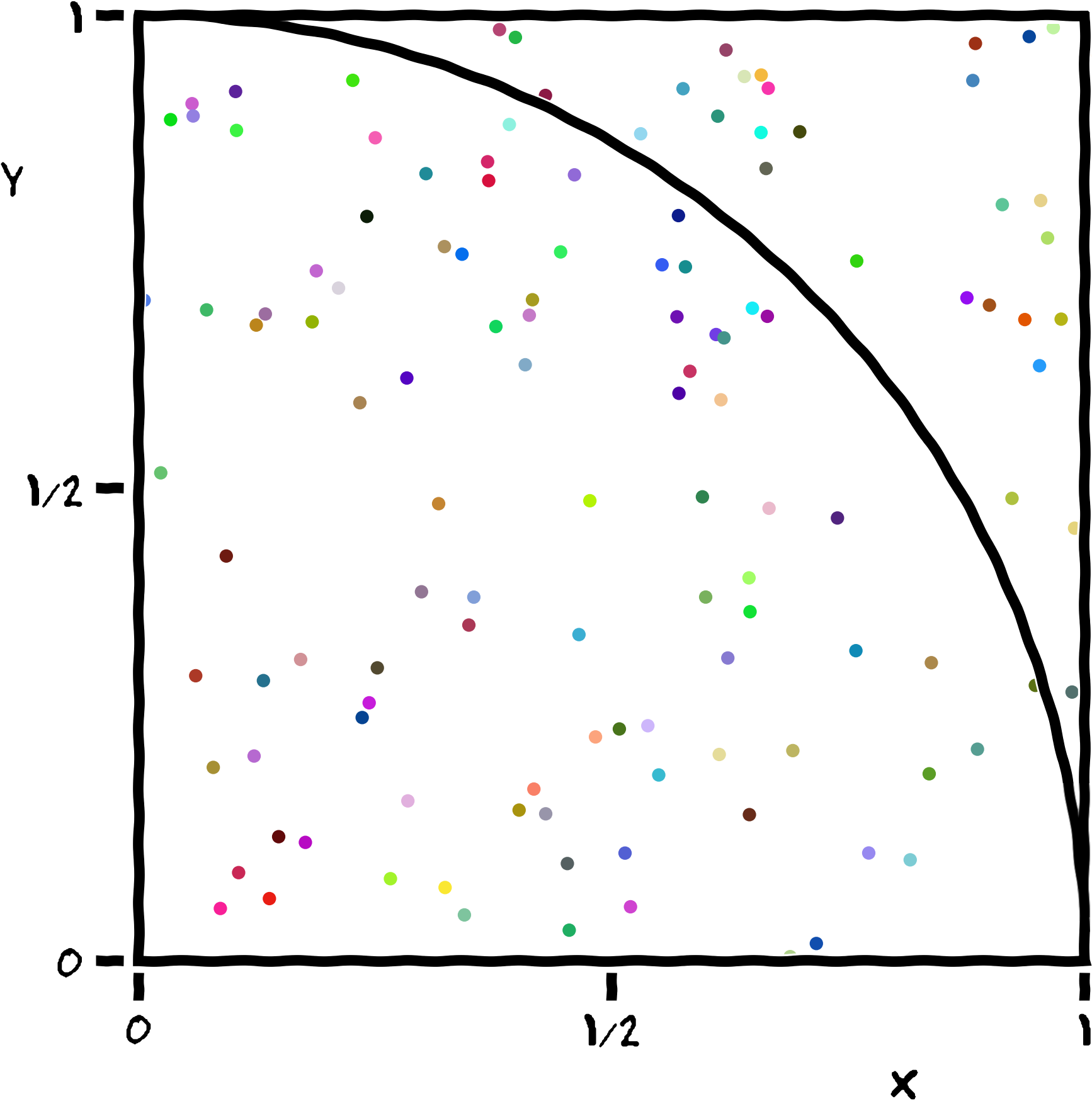
The program generates a large number of random points on a 1×1 square
centered on (½,½), and checks how many of these points fall inside the
unit circle. On average, π/4 of the randomly-selected points should fall
in the circle, so π can be estimated from 4f, where f
is the observed fraction of points that fall in the circle. Because each
sample is independent, this algorithm is easily implemented in
parallel.
Get code for this episode
The Python code you will use in this episode has been pre-written and
you can obtain a copy by: Method 1: Use the commands curl
or wget from the previous episode to download the files
directly into your working directory on Rocket and extract the archive.
Remember you will need to specify the path to these Python files in your
job submission scripts. It may be useful to cd into this
directory or mv the contents directly to the path
/nobackup/proj/training/userid.
BASH
[userid@rocket.hpc ~]$ curl -O https://nclrse-training.github.io/hpc-intro-cirrus/files/python-pi-code.tar.gz
[userid@rocket.hpc ~]$ tar -xvzf python-pi-code.tar.gzor
BASH
[userid@rocket.hpc ~]$ wget https://nclrse-training.github.io/hpc-intro-cirrus/files/python-pi-code.tar.gz
[userid@rocket.hpc ~]$ tar -xvzf python-pi-code.tar.gzMethod 2: You can download a local copy of the files on your machine
and then use scp or rsync to copy the file
onto Rocket.
A Serial Solution to the Problem
We start from a Python script using concepts taught in Software Carpentry’s Programming with Python workshops. We want to allow the user to specify how many random points should be used to calculate π through a command-line parameter. This script will only use a single CPU for its entire run, so it’s classified as a serial process.
Let’s write a Python program, pi.py, to estimate π for
us. Start by importing the numpy module for calculating the
results, and the sys module to process command-line
parameters:
We define a Python function inside_circle that accepts a
single parameter for the number of random points used to calculate π.
See Programming
with Python: Creating Functions for a review of Python functions. It
randomly samples points with both x and y on the
half-open interval [0, 1). It then computes their distances from the
origin (i.e., radii), and returns how many of those distances were less
than or equal to 1.0. All of this is done using vectors of
double-precision (64-bit) floating-point values.
PYTHON
def inside_circle(total_count):
x = np.random.uniform(size=total_count)
y = np.random.uniform(size=total_count)
radii = np.sqrt(x * x + y * y)
count = len(radii[np.where(radii<=1.0)])
return countNext, we create a main function to call the
inside_circle function and calculate π from its returned
result. See Programming
with Python: Command-Line Programs for a review of main
functions and parsing command-line parameters.
PYTHON
def main():
n_samples = int(sys.argv[1])
counts = inside_circle(n_samples)
my_pi = 4.0 * counts / n_samples
print(my_pi)
if __name__ == '__main__':
main()Run the code on your development machine (e.g. your laptop)
A normal workflow would be to develop and run scripts on your own
machine, then move the working code up to HPC if more resource is needed
to run it. If you have Python installed on your lapotop, feel free to
try this out: If we run the Python script locally with a command-line
parameter, as in python pi.py 1024, we should see the
script print its estimate of π:
Try out code on the login node ?
We only run small test jobs on the login node. Rather than have the whole class attempt this and block up the login node, your instructor will run the code as a demonstration: :::solution on Rocket, software is only available via modules, so we need to load Python3 before we start:
:::
Random Number Generation
Challenge
In the preceding code, random numbers are conveniently generated using the built-in capabilities of NumPy. In general, random-number generation is difficult to do well, it’s easy to accidentally introduce correlations into the generated sequence.
- Discuss why generating high quality random numbers might be difficult.
- Is the quality of random numbers generated sufficient for estimating π in this implementation?
- Computers are deterministic and produce pseudo random numbers using an algorithm. The choice of algorithm and its parameters determines how random the generated numbers are. Pseudo random number generation algorithms usually produce a sequence numbers taking the previous output as an input for generating the next number. At some point the sequence of pseudo random numbers will repeat, so care is required to make sure the repetition period is long and that the generated numbers have statistical properties similar to those of true random numbers.
- Yes.
Measuring Performance of the Serial Solution
The stochastic method used to estimate π should converge on the true
value as the number of random points increases. But as the number of
points increases, creating the variables x, y,
and radii requires more time and more memory. Eventually,
the memory required may exceed what’s available on our local laptop or
desktop, or the time required may be too long to meet a deadline. So
we’d like to take some measurements of how much memory and time the
script requires, and later take the same measurements after creating a
parallel version of the script to see the benefits of parallelizing the
calculations required.
Estimating Memory Requirements
Since the largest variables in the script are x,
y, and radii, each containing
n_samples points, we’ll modify the script to report their
total memory required. Each point in x, y, or
radii is stored as a NumPy float64, we can use
NumPy’s dtype
function to calculate the size of a float64.
Replace the print(my_pi) line with the following:
PYTHON
size_of_float = np.dtype(np.float64).itemsize
memory_required = 3 * n_samples * size_of_float / (1024**3)
print(f"Pi: {my_pi}, memory: {memory_required} GiB")The first line calculates the bytes of memory required for a single
64-bit floating point number using the dtype function. The
second line estimates the total amount of memory required to store three
variables containing n_samples float64 values,
converting the value into units of gibibytes.
The third line prints both the estimate of π and the estimated amount of
memory used by the script.
The updated Python script is:
PYTHON
import numpy as np
import sys
def inside_circle(total_count):
x = np.random.uniform(size=total_count)
y = np.random.uniform(size=total_count)
radii = np.sqrt(x * x + y * y)
count = len(radii[np.where(radii<=1.0)])
return count
def main():
n_samples = int(sys.argv[1])
counts = inside_circle(n_samples)
my_pi = 4.0 * counts / n_samples
size_of_float = np.dtype(np.float64).itemsize
memory_required = 3 * n_samples * size_of_float / (1024**3)
print(f"Pi: {my_pi}, memory: {memory_required} GiB")
if __name__ == '__main__':
main()Run the script again with a few different values for the number of samples, and see how the memory required changes:
BASH
[user@laptop ~]$ python pi.py 1000
Pi: 3.144, memory: 2.2351741790771484e-05 GiB
[user@laptop ~]$ python pi.py 2000
Pi: 3.18, memory: 4.470348358154297e-05 GiB
[user@laptop ~]$ python pi.py 1000000
Pi: 3.140944, memory: 0.022351741790771484 GiB
[user@laptop ~]$ python pi.py 100000000
Pi: 3.14182724, memory: 2.2351741790771484 GiBHere we can see that the estimated amount of memory required scales
linearly with the number of samples used. In practice, there is some
memory required for other parts of the script, but the x,
y, and radii variables are by far the largest
influence on the total amount of memory required.
Estimating Calculation Time
Most of the calculations required to estimate π are in the
inside_circle function:
- Generating
n_samplesrandom values forxandy. - Calculating
n_samplesvalues ofradiifromxandy. - Counting how many values in
radiiare under 1.0.
There’s also one multiplication operation and one division operation
required to convert the counts value to the final estimate
of π in the main function.
A simple way to measure the calculation time is to use Python’s
datetime module to store the computer’s current date and
time before and after the calculations, and calculate the difference
between those times.
To add the time measurement to the script, add the following line
below the import sys line:
Then, add the following line immediately above the line calculating
counts:
Add the following two lines immediately below the line calculating
counts:
And finally, modify the print statement with the
following:
The final Python script for the serial solution is:
PYTHON
import numpy as np
import sys
import datetime
def inside_circle(total_count):
x = np.random.uniform(size=total_count)
y = np.random.uniform(size=total_count)
radii = np.sqrt(x * x + y * y)
count = len(radii[np.where(radii<=1.0)])
return count
def main():
n_samples = int(sys.argv[1])
start_time = datetime.datetime.now()
counts = inside_circle(n_samples)
my_pi = 4.0 * counts / n_samples
end_time = datetime.datetime.now()
elapsed_time = (end_time - start_time).total_seconds()
size_of_float = np.dtype(np.float64).itemsize
memory_required = 3 * n_samples * size_of_float / (1024**3)
print(f"Pi: {my_pi}, memory: {memory_required} GiB, time: {elapsed_time} s")
if __name__ == '__main__':
main()Run the script again with a few different values for the number of samples, and see how the solution time changes:
PYTHON
[user@laptop ~]$ python pi.py 1000000
Pi: 3.139612, memory: 0.022351741790771484 GiB, time: 0.034872 s
[user@laptop ~]$ python pi.py 10000000
Pi: 3.1425492, memory: 0.22351741790771484 GiB, time: 0.351212 s
[user@laptop ~]$ python pi.py 100000000
Pi: 3.14146608, memory: 2.2351741790771484 GiB, time: 3.735195 sHere we can see that the amount of time required scales approximately linearly with the number of samples used. There could be some variation in additional runs of the script with the same number of samples, since the elapsed time is affected by other programs running on the computer at the same time. But if the script is the most computationally-intensive process running at the time, its calculations are the largest influence on the elapsed time.
Now that we’ve developed our initial script to estimate π, we can see that as we increase the number of samples:
- The estimate of π tends to become more accurate.
- The amount of memory required scales approximately linearly.
- The amount of time to calculate scales approximately linearly.
In general, achieving a better estimate of π requires a greater
number of points. Take a closer look at inside_circle:
should we expect to get high accuracy on a single machine?
Probably not. The function allocates three arrays of size N equal to the number of points belonging to this process. Using 64-bit floating point numbers, the memory footprint of these arrays can get quite large. Each 100,000,000 points sampled consumes 2.24 GiB of memory. Sampling 400,000,000 points consumes 8.94 GiB of memory, and if your machine has less RAM than that, it will grind to a halt. If you have 16 GiB installed, you won’t quite make it to 750,000,000 points.
Running the Serial Job on a Compute Node
Replicate the pi.py script in the
/nobackup/proj/training/userid space on Rocket. Guidance on
how to do this can be found at the beginning of this episode.
Create a submission file, requesting one task on a single node. If we
do not specify a maximum walltime for the job using
--time=<hh:mm:ss> then the job will be submitted with
the short default maximum time of 1 minute (NB partition
names and default time limits will vary between HPC systems).
OUTPUT
#!/bin/bash
#SBATCH --partition=short
#SBATCH --job-name serial-pi
#SBATCH --nodes=1
#SBATCH --tasks-per-node=1
# Load the correct Python module
module load python/3.9.13
# Execute the task
python3 pi.py 100000000Memory Requirements
On some HPC systems you may need to specify the memory requirements
of the job using the --mem, --mem-per-cpu,
--mem-per-gpu options. However, on Cirrus you cannot
specify the memory for a job. The amount of memory you are assigned is
calculated from the amount of primary resource you request.
The primary resource you request on standard compute nodes are CPU cores. The maximum amount of memory you are allocated is computed as the number of CPU cores you requested multiplied by 1/22 of the total memory available (as there are 22 CPU cores per node). So, if you request a full standard node (22 cores), then you will be allocated a maximum of all of the memory (128 GB) available on the node; however, if you request 1 core, then you will be assigned a maximum of 128/22 = 2.9 GB of the memory available on the node.e.
Then submit your job.
As before, use the status commands to check when your job runs. Use
ls to locate the output file, and examine it. Is it what
you expected?
- How good is the value for π?
- How much memory did it need?
- How long did the job take to run?
Modify the job script to increase both the number of samples (perhaps by a factor of 2, then by a factor of 10), and resubmit the job each time.
- How good is the value for π?
- How much memory did it need?
- Did you encounter any errors?
Even with sufficient memory for necessary variables, a script could require enormous amounts of time to calculate on a single CPU. To reduce the amount of time required, we need to modify the script to use multiple CPUs for the calculations. In the largest problem scales, we could use multiple CPUs in multiple compute nodes, distributing the memory requirements across all the nodes used to calculate the solution.
Running the Parallel Job
We will run an example that uses the Message Passing Interface (MPI) for parallelism – this is a common tool on HPC systems.
What is MPI?
The Message Passing Interface is a set of tools which allow multiple parallel jobs to communicate with each other. Typically, a single executable is run multiple times, possibly on different machines, and the MPI tools are used to inform each instance of the executable about how many instances there are, which instance it is. MPI also provides tools to allow communication and coordination between instances. An MPI instance typically has its own copy of all the local variables.
While MPI jobs can generally be run as stand-alone executables, in
order for them to run in parallel they must use an MPI run-time
system, which is a specific implementation of the MPI
standard. To do this, they should be started via a command such
as mpiexec (or mpirun, or srun,
etc. depending on the MPI run-time you need to use), which will ensure
that the appropriate run-time support for parallelism is included.
MPI Runtime Arguments
On their own, commands such as mpiexec can take many
arguments specifying how many machines will participate in the
execution, and you might need these if you would like to run an MPI
program on your laptop (for example). In the context of a queuing
system, however, it is frequently the case that we do not need to
specify this information as the MPI run-time will have been configured
to obtain it from the queuing system, by examining the environment
variables set when the job is launched.
What Changes Are Needed for an MPI Version of the π Calculator?
First, we need to import the MPI object from the Python
module mpi4py by adding an
from mpi4py import MPI line immediately below the
import datetime line.
Second, we need to modify the “main” function to perform the overhead and accounting work required to:
- subdivide the total number of points to be sampled,
- partition the total workload among the various parallel processors available,
- have each parallel process report the results of its workload back to the “rank 0” process, which does the final calculations and prints out the result.
The modifications to the serial script demonstrate four important concepts:
- COMM_WORLD: the default MPI Communicator, providing a channel for
all the processes involved in this
mpiexecto exchange information with one another. - Scatter: A collective operation in which an array of data on one MPI rank is divided up, with separate portions being sent out to the partner ranks. Each partner rank receives data from the matching index of the host array.
- Gather: The inverse of scatter. One rank populates a local array, with the array element at each index assigned the value provided by the corresponding partner rank – including the host’s own value.
- Conditional Output: since every rank is running the same
code, the partitioning, the final calculations, and the
printstatement are wrapped in a conditional so that only one rank performs these operations.
We add the lines:
immediately before the n_samples line to set up the MPI
environment for each process.
We replace the start_time and counts lines
with the lines:
PYTHON
if rank == 0:
start_time = datetime.datetime.now()
partitions = [ int(n_samples / cpus) ] * cpus
counts = [ int(0) ] * cpus
else:
partitions = None
counts = NoneThis ensures that only the rank 0 process measures times and
coordinates the work to be distributed to all the ranks, while the other
ranks get placeholder values for the partitions and
counts variables.
Immediately below these lines, let’s
- distribute the work among the ranks with MPI
scatter, - call the
inside_circlefunction so each rank can perform its share of the work, - collect each rank’s results into a
countsvariable on rank 0 using MPIgather.
by adding the following three lines:
PYTHON
partition_item = comm.scatter(partitions, root=0)
count_item = inside_circle(partition_item)
counts = comm.gather(count_item, root=0)Illustrations of these steps are shown below.
The Parallel Message Passing Interface (MPI) Process
Step 1: Setup the MPI environment and initialize local variables – including the vector containing the number of points to generate on each parallel processor:

Step 2: Distribute the number of points from the originating vector to all the parallel processors:

Step 3: Perform the computation in parallel:

Step 4: Retrieve counts from all the parallel processes:

Step 5: Print out the report:

Finally, we’ll ensure the my_pi through
print lines only run on rank 0. Otherwise, every parallel
processor will print its local value, and the report will become
hopelessly garbled:
if rank == 0:
my_pi = 4.0 * sum(counts) / sum(partitions)
end_time = datetime.datetime.now()
elapsed_time = (end_time - start_time).total_seconds()
size_of_float = np.dtype(np.float64).itemsize
memory_required = 3 * sum(partitions) * size_of_float / (1024**3)
pi_specific = np.pi
accuracy = 100*(1-my_pi/pi_specific)
print(f"Pi: {my_pi:6f}, memory: {memory_required:6f} GiB, time: {elapsed_time:6f} s, error: {accuracy:6f}%")A fully commented version of the final MPI parallel python code is available: pi-mpi-cirrus.py.
Our purpose here is to exercise the parallel workflow of the cluster, not to optimize the program to minimize its memory footprint. Rather than push our local machines to the breaking point (or, worse, the login node), let’s give it to a cluster node with more resources.
Create a submission file, requesting more than one task on a single node:
OUTPUT
#!/bin/bash
#SBATCH --partition=short
#SBATCH --job-name parallel-pi
#SBATCH --nodes=1
#SBATCH --tasks-per-node=4
#SBATCH --time=00:01
# Load the correct Python module
module load python/3.9.13
# Execute the task
srun python pi-mpi-cirrus.py 100000000Then submit your job.
As before, use the status commands to check when your job runs. Use
ls to locate the output file, and examine it. Is it what
you expected?
- How good is the value for π?
- How much memory did it need?
- How much faster was this run than the serial run with 100000000 points?
Modify the job script to increase the number of samples (perhaps by a factor of 2, then by a factor of 10), and resubmit the job each time. You can also increase the number of CPUs.
- How good is the value for π?
- How much memory did it need?
- How long did the job take to run?
How Much Does MPI Improve Performance?
In theory, by dividing up the π calculations among n MPI processes, we should see run times reduce by a factor of n. In practice, some time is required to start the additional MPI processes, for the MPI processes to communicate and coordinate, and some types of calculations may only be able to run effectively on a single CPU.
Additionally, if the MPI processes operate on different physical CPUs in the computer, or across multiple compute nodes, additional time is required for communication compared to all processes operating on a single CPU.
Amdahl’s Law is one way of predicting improvements in execution time for a fixed parallel workload. If a workload needs 20 hours to complete on a single core, and one hour of that time is spent on tasks that cannot be parallelized, only the remaining 19 hours could be parallelized. Even if an infinite number of cores were used for the parallel parts of the workload, the total run time cannot be less than one hour.
In practice, it’s common to evaluate the parallelism of an MPI program by
- running the program across a range of CPU counts,
- recording the execution time on each run,
- comparing each execution time to the time when using a single CPU.
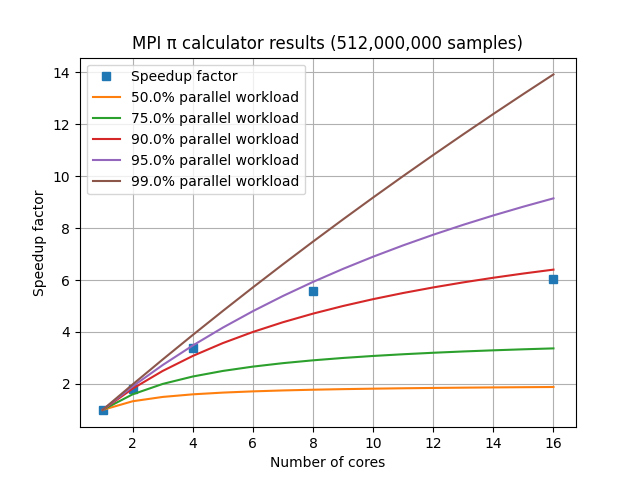
The speedup factor S is calculated as the single-CPU execution time divided by the multi-CPU execution time. For a laptop with 8 cores, the graph of speedup factor versus number of cores used shows relatively consistent improvement when using 2, 4, or 8 cores, but using additional cores shows a diminishing return.
Laptop performance
For a set of HPC nodes containing 28 cores each, the graph of speedup factor versus number of cores shows consistent improvements up through three nodes and 84 cores, but worse performance when adding a fourth node with an additional 28 cores. This is due to the amount of communication and coordination required among the MPI processes requiring more time than is gained by reducing the amount of work each MPI process has to complete. This communication overhead is not included in Amdahl’s Law.
HPC performance
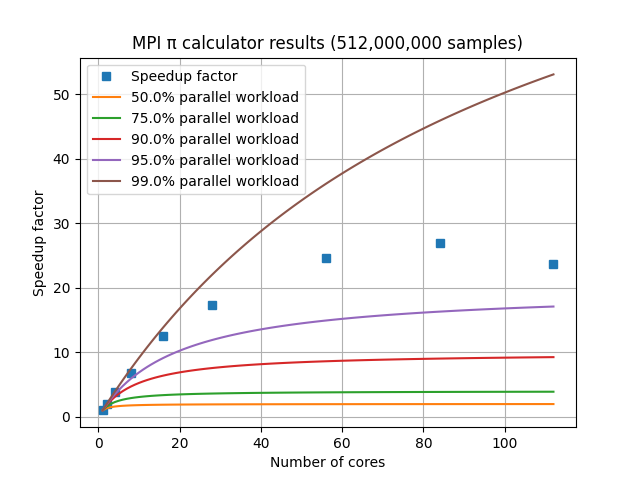
In practice, MPI speedup factors are influenced by:
- CPU design,
- the communication network between compute nodes,
- the MPI library implementations, and
- the details of the MPI program itself.
In an HPC environment, we try to reduce the execution time for all types of jobs, and MPI is an extremely common way to combine dozens, hundreds, or thousands of CPUs into solving a single problem. To learn more about parallelization, see the parallel novice lesson lesson.
- “Parallel programming allows applications to take advantage of parallel hardware; serial code will not ‘just work.’”
- “Distributed memory parallelism is a common case, using the Message Passing Interface (MPI).”
- “The queuing system facilitates executing parallel tasks.”
- “Performance improvements from parallel execution do not scale linearly.”
